Biotechnology is a broad and relatively new field that studies living organisms’ biological processes or systems to harness them in producing useful products. These products cover broad applications and industries including agricultural, pharmaceutical, medical, and environmental. The main purpose of this field is to use what exists in nature to create more efficient machines, and to solve problems using biology. One of the key applications of biotechnology is the direct manipulation of an organism’s genome, known as genetic engineering.
The Discovery of DNA
The road to genetic engineering starts with Watson and Crick, and some form of injustice. In 1963, James Watson and Francis Crick won the Nobel prize for discovering the double helix structure of deoxyribonucleic acid (DNA), but another DNA researcher named Rosalind Franklin should have been recognized alongside this duo. Rosalind Franklin earned her doctorate in 1945 and went on to become an expert at X-ray diffraction photography. This method allowed her to photograph structures that had never before been seen.
By using X-ray diffraction photography, Franklin became the first person to see and photograph DNA. A major part of science is the sharing of research and discoveries and Franklin did just that by showing her photograph of DNA to the scientific community. This photograph is titled Photo 51, and it was the image Watson and Crick used to determine the structure of DNA. This in itself is not inappropriate because scientists build off of each other’s work all the time. However, Watson and Crick used Franklin’s photographs without her permission and ended up taking credit for the discovery in its entirety, owing to the fact that they submitted their research for publishing a day before Franklin did.
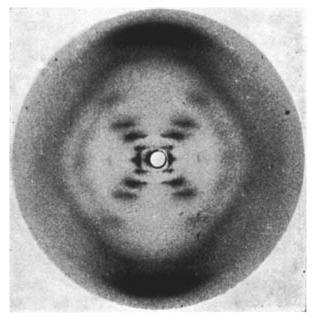
Nonetheless, the discovery of the double helix structure of DNA has helped explain the longevity of DNA and the mechanism of genetic inheritance. Many leaps were made in the scientific understanding of DNA thanks to Franklin, Watson, and Crick’s research, but the discovery of recombinant DNA, or rDNA by Cohen and Boyer in the 1970s would be the one to contribute to the field of genetic engineering specifically. rDNA is synthetic DNA that is made by inserting sections from the genome of one organism into the genome of another. This invention enabled new avenues of research regarding the study of specific genes, but it also ignited a storm of ethical questions surrounding genetic editing. At the same time, the potential of using this technology for curing diseases made it invaluable.
Early Genetic Engineering: Synthetic Insulin
About a decade after the creation of rDNA, one of the most notable discoveries in the field was made based on this technology. Researchers found a way to produce synthetic insulin or ‘human insulin’. Patients with type-1 diabetes used to take insulin derived from animals, usually that of pigs and cows. Synthetic insulin was produced by rDNA technology, where the gene encoding human insulin was inserted into a plasmid, bacterial DNA which exists in a continuous loop rather than as a string. This plasmid would then be introduced into a host bacterium, which would multiply, producing the recombinant bacteria. The recombinant bacteria would then produce human insulin as it divided due to the presence of the human insulin gene in its plasmid. Following this, the insulin would be purified from the bacterial culture.
On the heels of this discovery was the first recombinant Hepatitis B vaccine, and the start of genetically modified crops (GMOs). The first of these crops is called Bt corn, as it contained genes from the bacteria Bacillus thuringiensis (Bt). The cloning of Dolly the sheep and whole-genome sequencing followed suit.
The Creation of CRISPR, and the Future of Genetic Engineering
In 2020 two scientists, Emmanuelle Charpentier and Jennifer Doudna won the Nobel Prize in Chemistry for the discovery of Clustered Regularly Interspaced Short Palindromic Repeats, more commonly referred to as CRISPR. CRISPR was first identified around 2011 as a component of the bacterial immune system. Bacteria are single-celled organisms, and so their immune systems are quite different from our own, but they still need it to protect against viruses, or phages as they are referred to in bacteria. A virus is able to identify a host cell (human or otherwise) by receptors on the cell’s surface. Once this happens, the virus will attach itself to the cell and inject its viral DNA. If the virus is successful in infecting the host cell, the viral DNA will act as instructions that force the cell to make more of the virus.
The bacterial immune system is able to identify viral DNA and take it apart, using CRISPR. The CRISPR system has two main parts: a strand of RNA known as guide(g)RNA, and a Cas9 protein. The gRNA is similar to a single, or unmatched strand of DNA, and it is this part of the complex that can match with and identify viral DNA. A Cas9 (CRISPR associated 9) protein is attached to the gRNA, and once the gRNA binds to the viral DNA, the Cas protein will cut it up into pieces, destroying it. This complex is cumulatively referred to as CRISPR Cas9.
However, the Nobel Prize winners, Charpentier and Doudna determined that CRISPR Cas9 could be placed into any cell and cut any DNA targeted by the gRNA. Scientists are able to design gRNA to match any DNA sequence, and so with this discovery, the possibilities for genetic engineering and biotechnology have become near-infinite. Now, we have immense development and progress with innovations taking place every single day! It is undoubtedly an incredible time to be working in the wondrous and ever-growing field of biotechnology!
Enjoyed this article? Visit the 4P Academy Blog for more informative pieces on a variety of fascinating topics.
SOURCES
Elkin, O. Rosalind Franklin, and the double helix. Physics Today. 2003 56(3), 42-48.
Berg, P., & Mertz, E. Personal reflections on the origins and emergence of recombinant DNA technology. Genetics. 2010184(1), 9-17.
Vitetta, E. S., Berton, M. T., Burger, C., Kepron, M., Lee, W. T., & Yin, X. M. (1991). Memory B and T cells. Annual review of immunology, 9(1), 193-217.
Groth, S. & Scheidegger, D. Production of Monoclonal Antibodies: Strategy and Tactics. Journal of Immunological Methods. 1980. 35(1-2), 1-21.
Ledford, H., & Callaway, E. Pioneers of Revolutionary CRISPR Gene editing Win Chemistry Nobel. Nature News. 2020. https://www.nature.com/articles/d41586-020-02765-9
Tibaldi, M. Evolution of insulin development: focus on key parameters. Advances in Therapy. 2012. 29(7), 590-619.
Watson, J ., Watson, C., & Gilman, M., et al. Recombinant DNA. Macmillan.1992
Douville, M., Gagné, F., & Blaise, C., et al. Occurrence and Persistence of Bacillus Thuringiensis (Bt) and Transgenic Bt corn cry1Ab Gene from an Aquatic Environment. Ecotoxicology and Environmental Safety. 2007. 66(2), 195-203.

